QA4IQI: Quality Assessment for Interoperable Quantitative CT-Imaging 
Medical images form one of the corner stones of diagnosis for various conditions. They provide not only non-invasive visual observations but also allow quantitative measurements of size and tissue texture. Similar to most measurement devices, such as a simple ruler, medical imaging devices also need to be calibrated. While calibration for geometric measurements and tissue density are commonly performed, features that quantify more complex characteristics are often neglected. For example, tissue heterogeneity is correlated with aggressiveness in various cancer types. Quantitative metrics that measure heterogeneity are not currently considered during calibration, which makes medical imaging difficult to use as a measurement tool. This project aims to close this gap.
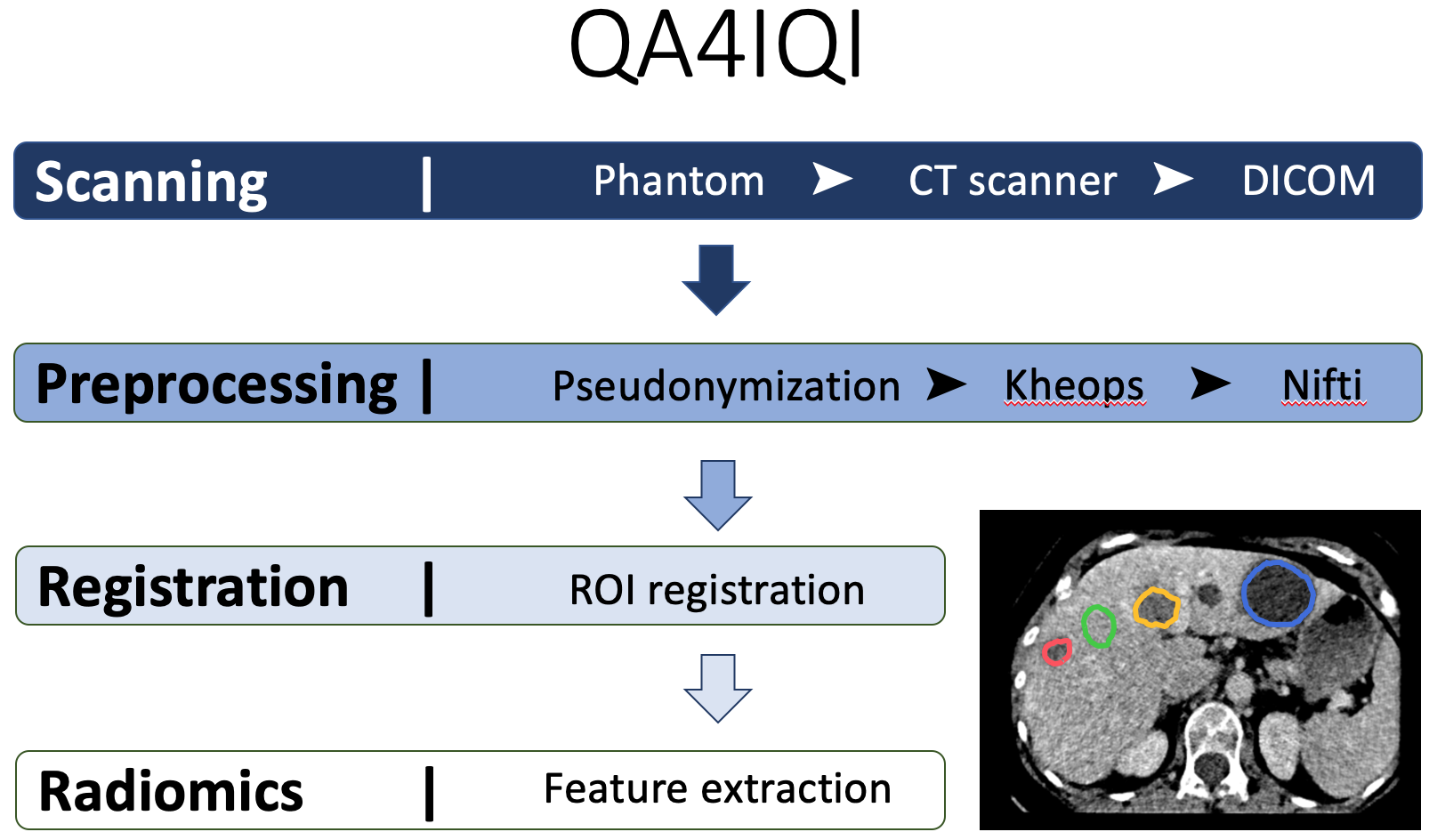
The QA4IQI project aims to build infrastructure crucial for integrating imaging data in research in personalised health and translating advanced image computing techniques to clinical practice. It will aid the establishment of both an automated imaging quality assessment and quantitative parameter extraction pipeline. In this fashion, unstructured imaging data can automatically be transformed in structured, imaging-derived quantitative parameters.
The proposed approach to assess the compromise between feature stability and discriminative power is summarized in the following paper: Jimenez del Toro et al., “The discriminative power and stability of radiomics features with CT variations: Task-based analysis in an anthropomorphic 3D-printed CT phantom”, Investigative Radiology, In press.
When compared to other approaches assessing the influence of CT parameters on radiomics features, the advantages of our approach lies the two following aspects:
- The use of a 3D-printed radio-opaque and anthropomorphic phantom allowing repeated multiple scans without irradiating a real patient. This yields high statistical power to assess both feature stability and discriminatory power.
- The assessment of the discriminative power of features, allowing to put into perspective the intra-class variability resulting from feature instabilities.
Impact of reconstruction kernel on image appearance:
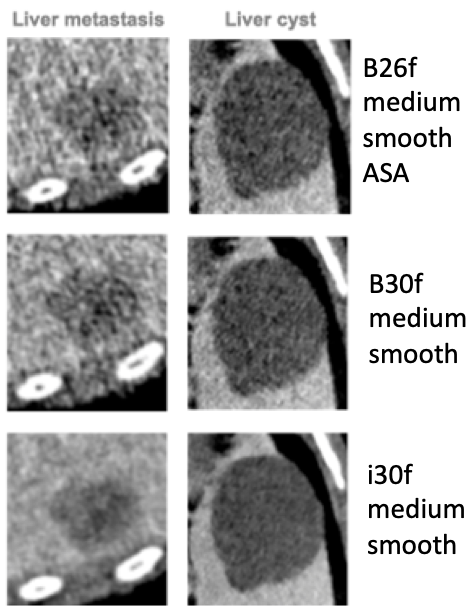
The phantom and regions of interests used to measure stability and and discriminatory power in the liver. They include 2 normal liver tissue areas (green), 2 benign cysts (blue), 1 liver metastasis from a colon carcinoma (red) and 1 hemangioma (yellow).
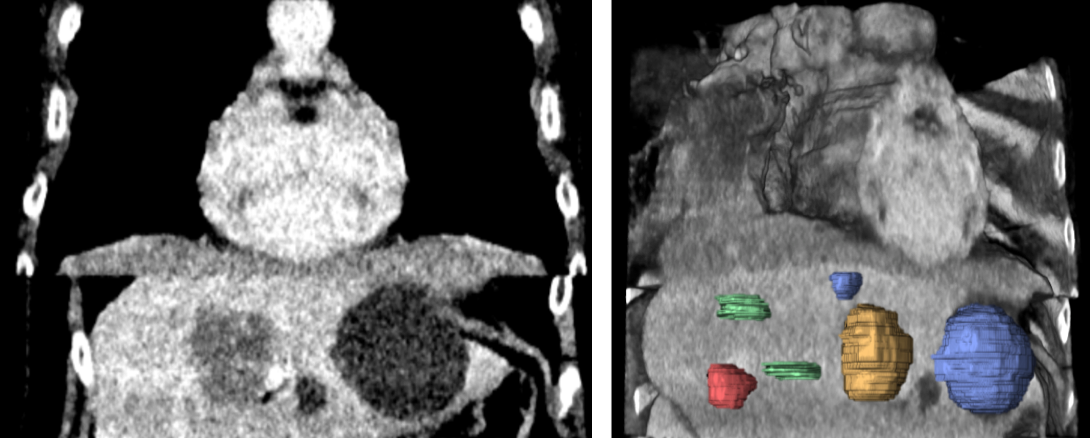
A principal component overview of intraclass variability seen in a feature space spanned by 86 radiomics features, populated with 240 CTs from 30 repeated phantom acquisitions for each of the 8 groups of CT recon. parameter variation (2 recon. algorithms, 4 recon. kernels, 4 slice thicknesses, 3 slice spacings):
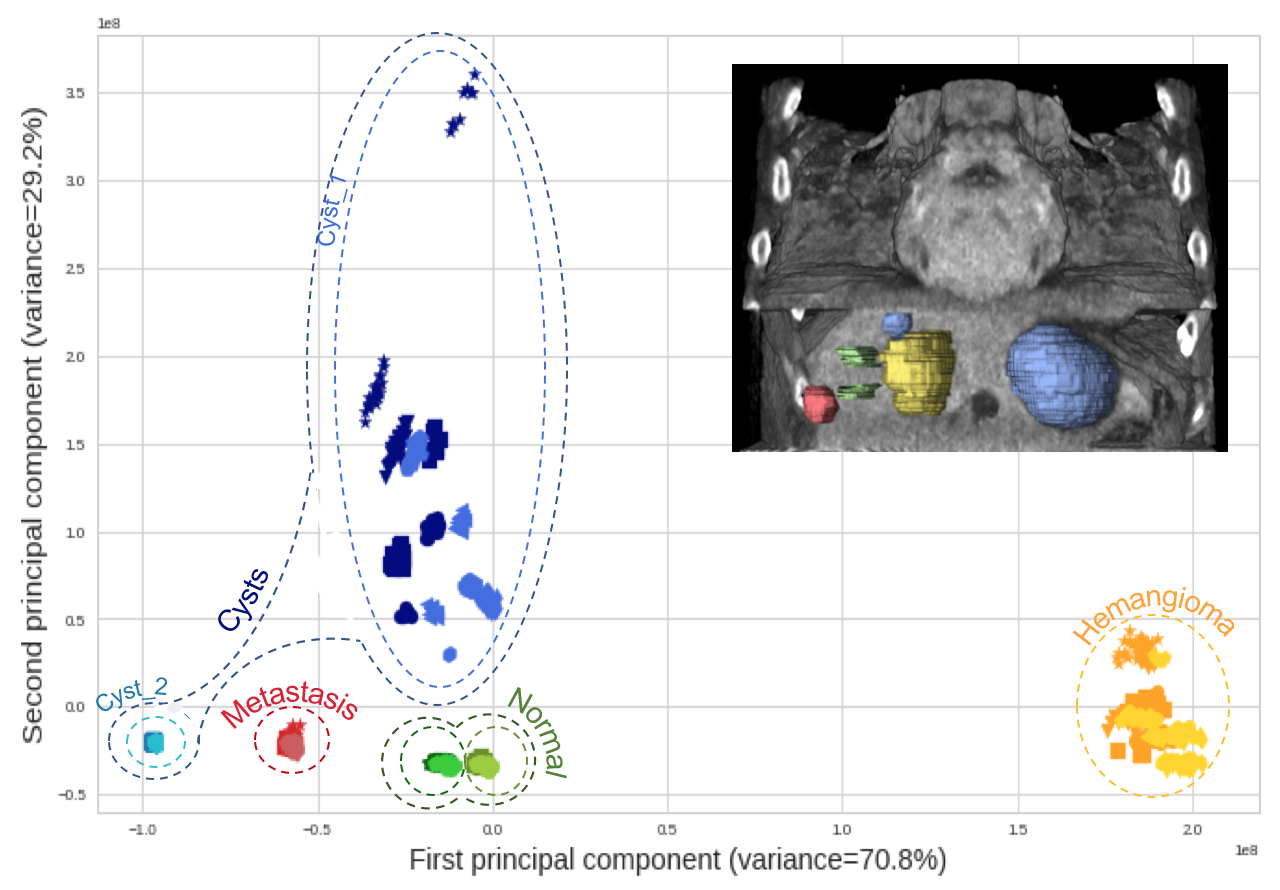
This reveals that the intraclass variability caused by CT parameters remains limited when compared to inter-class variability.
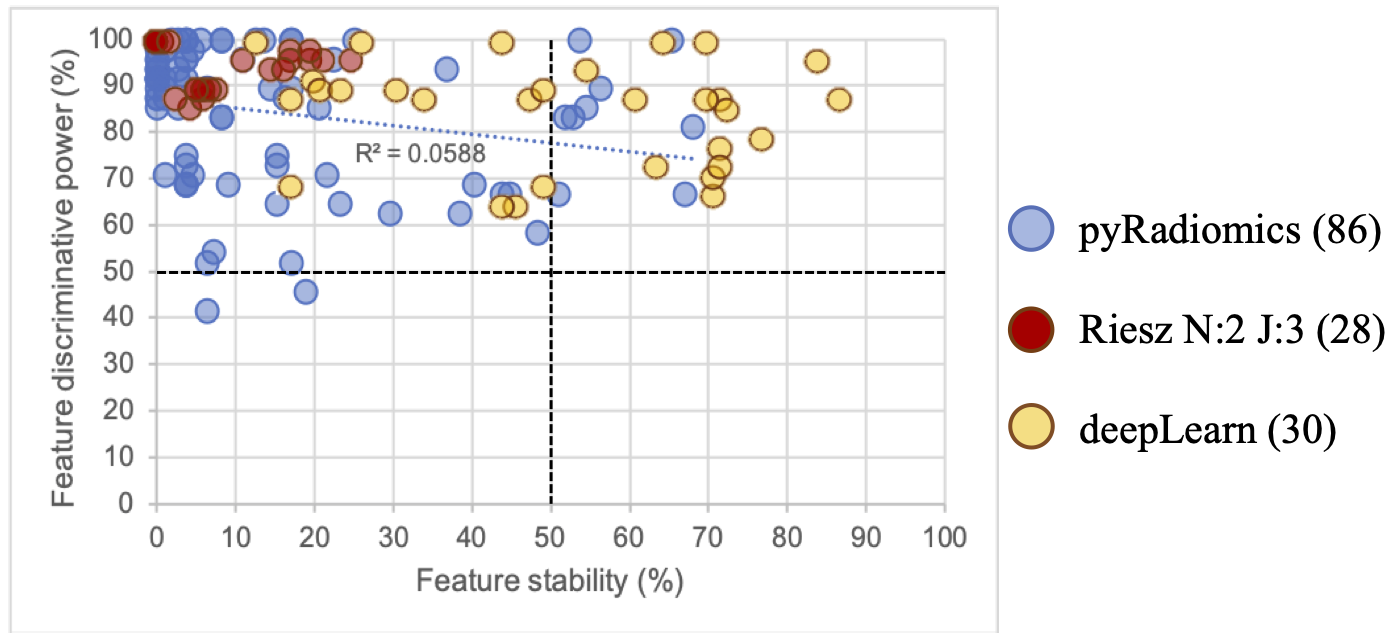
The proposed analysis comparing the discriminative power and stability of various features types reveals that deep features seem to achieve the best trade-off in this context.
The project is supported by the Swiss Personalized Health Network (SPHN).
Datasets
A first public dataset is available on TCIA, please refer to the following link for a description of the dataset and download instructions : https://doi.org/10.7937/a1v1-rc66
Feature Extraction Code & Docker Container
The following repository contains the source code & instructions on how to run the full feature extraction pipeline on the TCIA dataset : https://github.com/QA4IQI/qa4iqi-extraction






